Appendix C — Convert DMCs table to BED
This section describes how to convert the DMCs table to standard BED format using the ChAMP2bed.py script.
C.1 Method
Python3 must be installed on your system, no additional libraries are required. If your system lacks any python installation, please refer to this page: Python 3 Installation & Setup Guide.
C.1.1 Description
To use ChAMP2bed.py open a terminal and move to the directory storing your main methylR results. ChAMP2bed.py must be in the same directory storing your DMCs table:
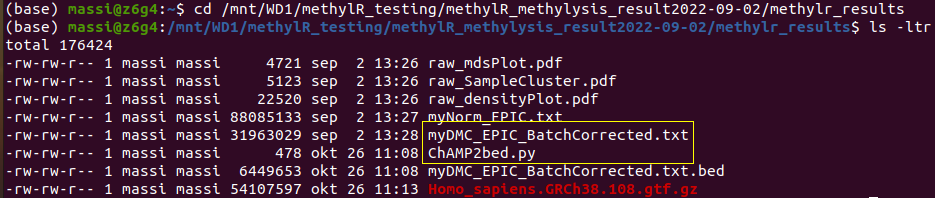
Run the command:
python3 ChAMP2bed.py myDMC_EPIC_BatchCorrected.txtOr adjust with the actual filename for your table. It will produce a new file with the same filename from your table but with the .bed extension:
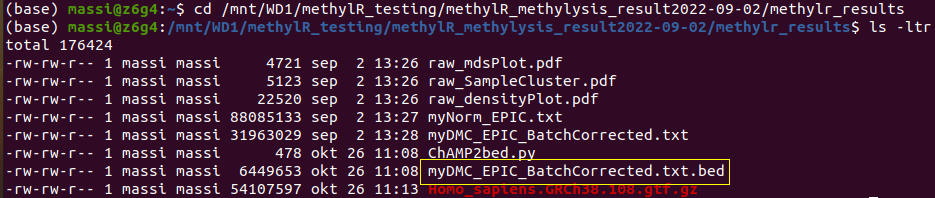
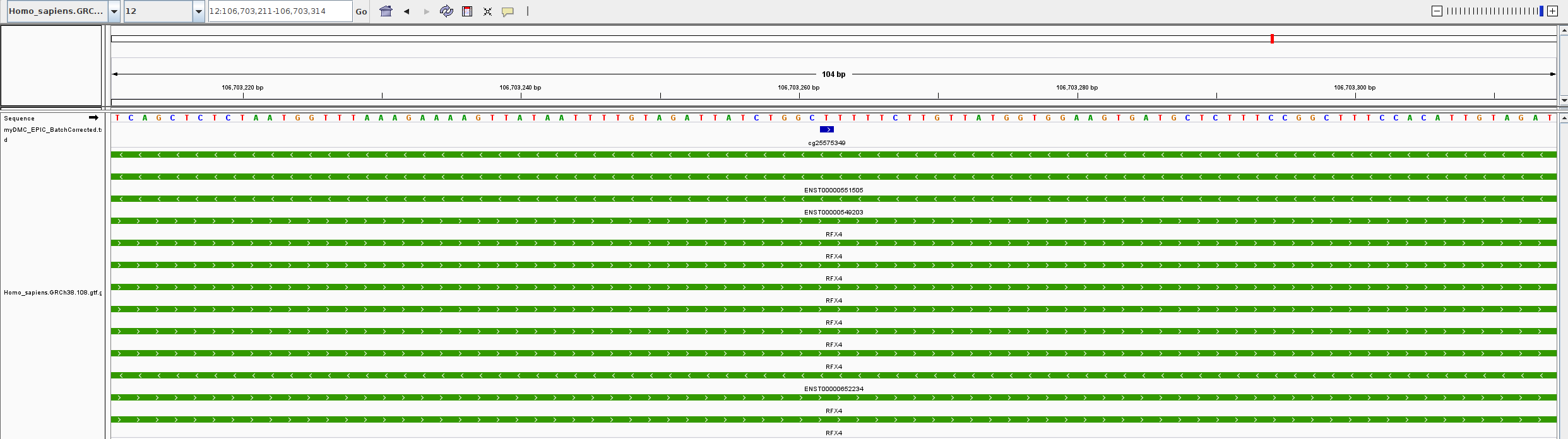
You can use this file as input for Gviz or import it either in IGV or as a custom track in any other genome browser. Be sure to match the proper genome version used to perform the analysis and to download the correct GTF/GFF3 file if you want to display the CpG (“blue”) together with additional features, such as genes (“green”):
Copyright © 2022-2023. Massimiliano Volpe and Jyotirmoy Das.
